Overview
The 3D Brain Atlas Analysis Tool is designed for automatic segmentation of brain image data. Automated fusion routines register an application-specific brain atlas to pre-clinical image data (or vice versa) and advanced statistical analysis methods are employed to provide rapid and thorough region-specific volume and uptake quantification.
Use the 3D Brain Atlas Tool to perform brain region analysis. The tool can handle CT or MR data as the input and MR, PET, or SPECT for the input functional data.
Install atlas
Please download and install the available atlases by going to Advanced Modules > Install Atlas > Mouse & Rat.
To change where the atlases are automatically stored, set via Advanced Modules > Install Atlas > Change Atlas directory option.
Typically the default path is located next to the VivoQuant cache:
| Platform | Default path |
|---|---|
| macOS | ~/Library/VivoQuant/atlas |
| Ubuntu | ~/.VivoQuant/atlas |
| Windows | %appdata%/Perceptive/VivoQuant/atlas |
For information on creating an atlas please refer to the How to create a Custom Brain Atlas page.
Prepare Image Data
Please load an anatomical image (CT or MR) and, if applicable, its corresponding NM data set before proceeding.
The tool requires the anatomical image from the data set to be loaded in the reference position. Any subsequent input images must be co-registered to the reference image prior to using the tool.
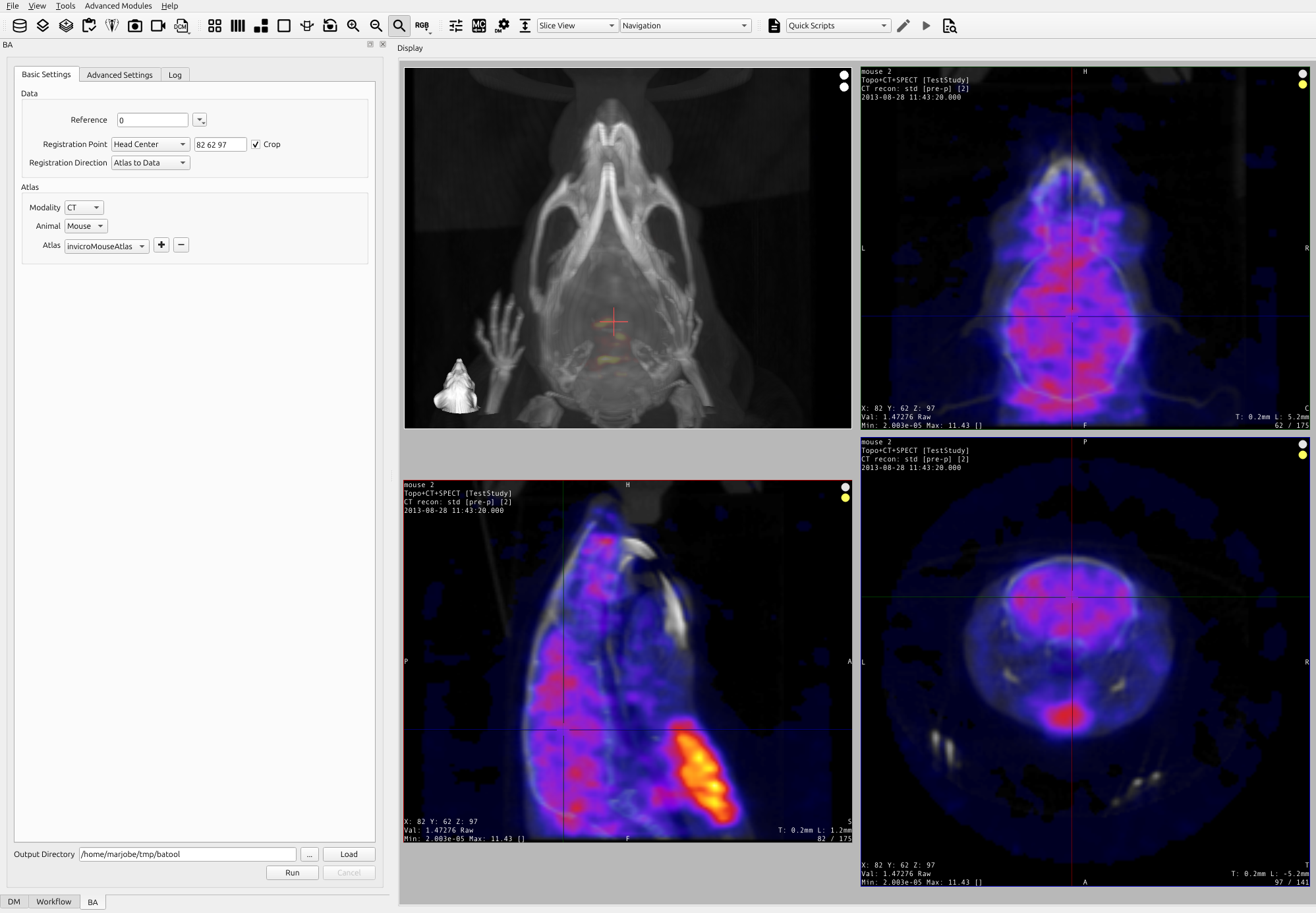
Providing a seed point is necessary if the images are not focused around the head/brain. To provide a seed point please place the cursor in the center of the brain in the Sagittal, Coronal, and Transverse views as seen in the image below.
Prepare MR Image Data
When using an MR data set as the reference image, a more accurate registration of the atlas can be achieved by performing an additional pre-processing step. Skull stripping the MR can be done semi-automatically using the 3D ROI Tool.
Segment Brain ROI
The ‘Spline Tool’ tab in the 3D ROI tool has an automatic thresholding tool which will allow the user to automatically segment the brain by providing a seed point on each brain slice. If necessary, the user can adjust the threshold for each individual slice to edit the automatic segmentation. To apply the segmentation for a given slice, use the green check mark button. To apply the segmentation and move on to the next slice, use the green check mark button with the red plus sign.
Cut Background ROI
Once satisfied with the brain segmentation on each slice, the user should cut the background ROI to complete the skull stripping. To cut the background ROI, set the background ROI as the output ROI and click on the ‘Cut ROI’ icon (circled in red in the image below) under the ‘Navigation’ tab in the 3D ROI Tool.
Result
All voxels that were not in the segmented brain ROI will be cut out of the image.
3D Brain Atlas Tool Settings GUI: Basic Settings
Getting There
To open the Atlas Tool settings GUI, please go to Advanced Modules > 3D Brain Atlas Tool
Basic Settings Tab
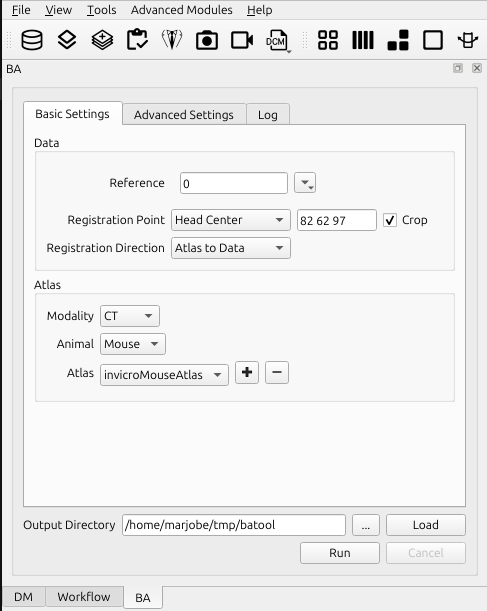
Data
Reference
Choose the reference image to perform the brain atlas
Registration Point
Select the registration point.
- Choose “Head Center”: will center the reference’s head and do all the operations to present the brain atlas, it is the best method for cropped output. Options ‘Registration Direction’ and ‘Crop’ are enabled for this technique.
- Choose “Cursor Seed Point”: will seed the operation with the crosshair point in order to present the brain atlas, it is the best method for non-cropped output. In this technique ‘Registration Direction’ is “Atlas to Data” and ‘Crop’ is unchecked, both always fixed.
Registration Direction
Select the registration direction.
- Choose “Data to atlas” to transform the image data (including anisotropic scaling and shearing) to match the atlas.
- Choose “Atlas to data” to transform the atlas to match the anatomical reference image.
Crop
Check the crop box if the data are not focused. The current head center (or seed point) will be displayed. To update the head center with the tool window open, use the mouse scroll wheel to move the crosshairs.
Atlas
Modality
Select the modality of the anatomical reference image. We currently support CT and MR images. If the image is tagged with modality information, the tool should detect this automatically.
Animal
Select the appropriate animal type.
- For CT registration, we currently support mouse, rat, squirrel monkey, and cynomolgus monkey.
- For MR registration, we currently support mouse, rat, canine, cynomolgus monkey, and rhesus monkey.
Atlas
Select the appropriate atlas type. These are populated based on the modality, animal type, and contents of the user’s atlas directory.
For information on creating an atlas please refer to the How to create a Custom Brain Atlas page.
Output Directory
Select the output directory in which to store results. If there are existing results in this directory, they will be deleted when the tool is run. To load the existing results click the Load button. The following will be stored here:
| File | Purpose |
|---|---|
| atlasFinal.rhma | The output brain ROI |
| atlasFinal-anatomical.mhd | Anatomical reference used for registration |
| data.csv | Uptake and concentration data for each brain region for each input frame |
| data-montage.png | QC image of ROI fitted to data |
| settings.txt | Settings used to generate output |
| tmp-XXX.mhd | Output scans after transforms, starting with XXX = 000 |
| tmp-XXX.raw | Raw file of associated with the pairing mhd file |
Run the Tool
Click the Run button to begin processing. The dialog window will switch to the “Log” tab where progress updates are displayed.
- Click the Cancel button to attempt to abort processing. This will stop processing after the current stage of processing has finished. As registration accounts for most of the processing time, this may not be responsive.
- When the tool finishes, results will load automatically (including the CSV file) and VQ will finish in the 3D ROI tool.
- Processing time will depend upon the computer’s inherent capabilities and the file size of the image files.
3D Brain Atlas Tool Settings GUI: Advanced Settings
Advanced Settings Tab
The advanced settings tab allows settings to be controlled that normally will not need to be modified.
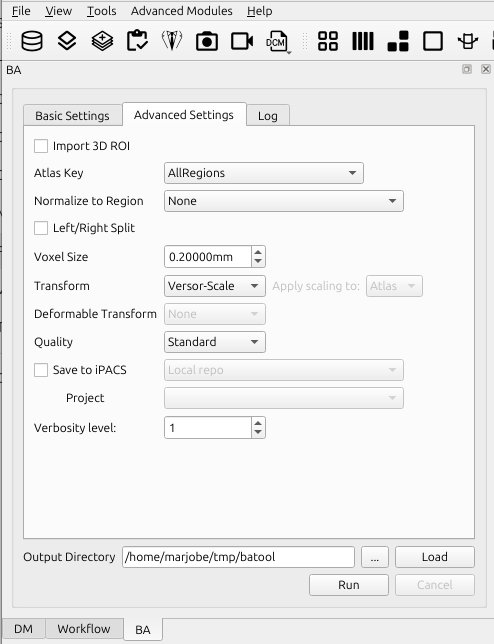
Import 3D ROI
Check this box to use the ROIs currently in the 3D ROI tool instead of a specific brain atlas. If this box is checked, no cropping registration will be performed, and a new output spreadsheet will be generated. This is typically enabled (checked) when the tool has already been run before and ROIs have already been fitted and need to be adjusted, or when a .rmha file is provided to you by the analysis lead and you are required to manually fit the ROIs.
Atlas Key
The atlas key controls how the original atlas regions are mapped to the final atlas.
- AllRegions (default): Combines left/right regions.
- AllRegionsLeft: Retains left regions only.
- AllRegionsRight: Retains right regions only.
- AllRegionsLeftRight: Retains left and right regions.
Normalize to Region
Select an ROI from the drop down menu to normalize concentrations in the spreadsheet to that region.
Left/Right Split
Check this box to output both the original atlas and the split atlas. This only works if there are “Left” and “Right” keys for the selected atlas key. For example, “AllRegions” will work if “AllRegionsLeft” and “AllRegionsRight” exist.
Voxel Size
Change the voxel size of the output data if a different resolution from the input data is desired.
- Lower values (higher resolution) are more computationally expensive.
- Higher values (lower resolution) will speed processing but may reduce atlas ROIs to be unreasonably small.
- Defaults to use same size as the starting image
Transform
Change the transform to restrict the number of transform parameters.
- Versor: Rigid body transform, translation and rotation only.
- Versor-Scale: Rigid body transform followed by scaling of the atlas (regardless of the registration option from basic settings tab).
- Versor-Affine: Affine transform followed by minor scaling/shearing.
- Affine: Affine transform only (faster but less robust than Versor-Affine).
Deformable Transform
Only for MR modality, the user can choose a deformable transform to use. Unlike rigid or affine transformations, a deformable transformation allows for the individual parts of the brain to be moved and warped independently.
- None: no deformable transform is applied.
- LogDemons: essentially a local displacement vector that moves a point in the moving image closer to a corresponding point in the fixed image, the update is then smoothed to ensure a smooth deformation.
- SyN: Symmetric Normalization is a highly popular and effective deformable registration algorithm, its defining feature is its symmetric nature.
Quality
Change the quality to improve registration results and/or processing speed.
- Fast: faster than standard, but less robust.
- Standard (default).
- Fine: slower than standard, but more robust (e.g. ideal for large rotations).
Save to iPACS
Check this box to upload images and ROIs directly to an iPACS.
- Only iPACS servers that have been previously connected to via open DICOM will be listed.
- Results will also be saved locally in the “Output Directory”.
Verbosity level
Change the level from 0 to 3, higher the value the higher the level of verbosity.
Manual Correction
Sometimes automatic registration will be poor and will need manual correction.
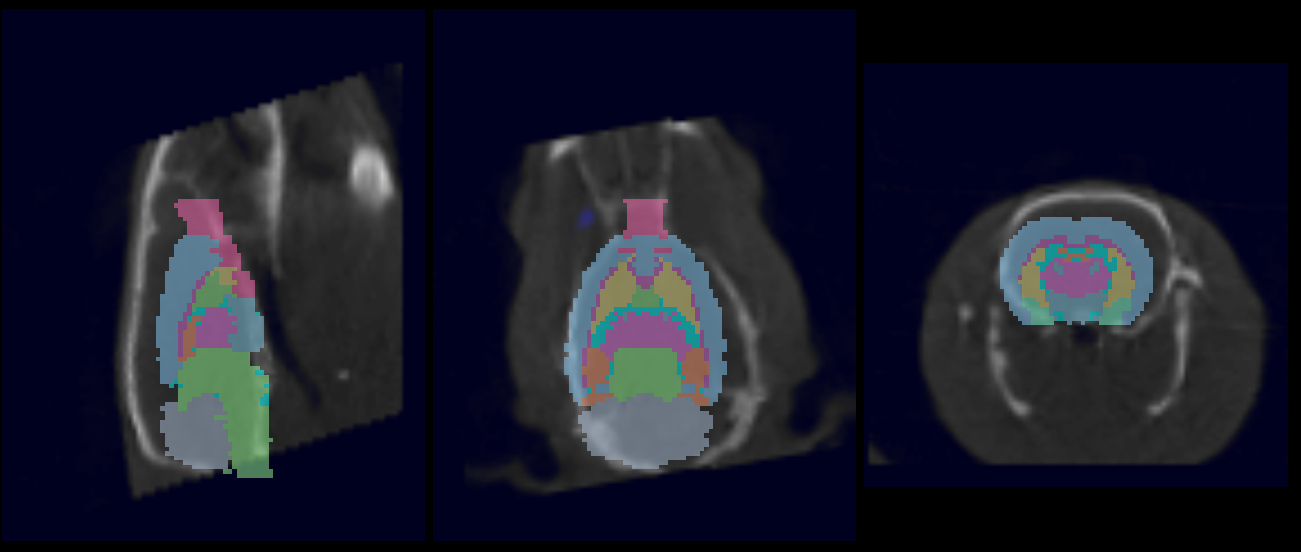
To manually edit the registration, load the ‘atlasFinal-anatomical.mhd’ file from the output directory and place it in the reference position. Navigate to the Reorientation/Registration operator and manually correct the registration:
- Data to Atlas (seen in example image below): Register all inputs to the atlas anatomical reference.
- Atlas to Data: Register the atlas anatomical reference to the input anatomical. Apply this transform to the ROI.
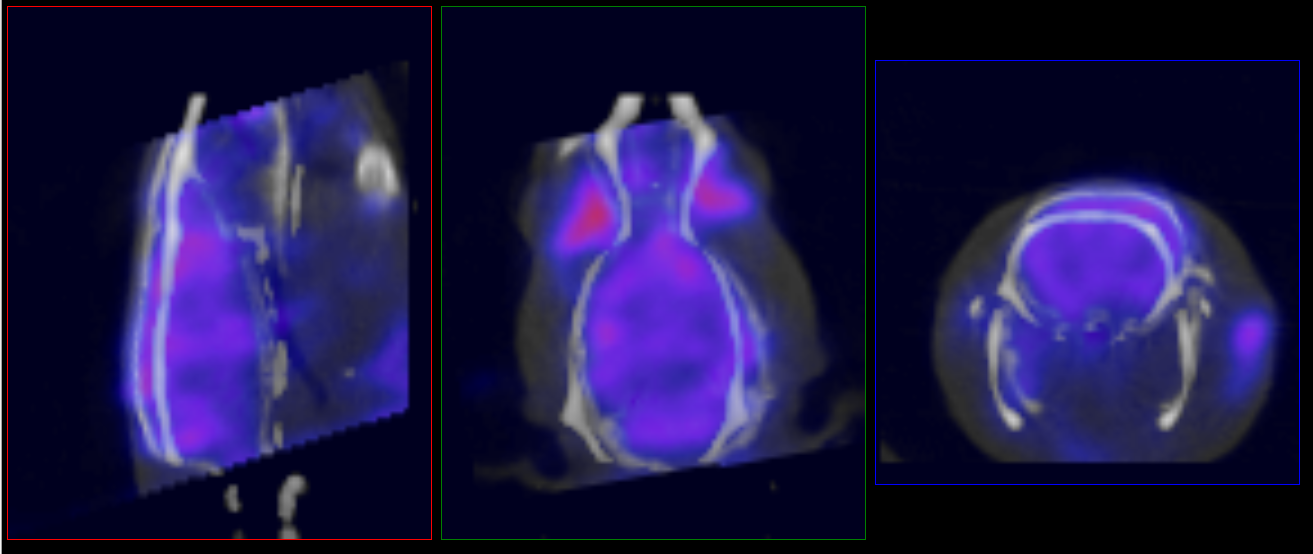
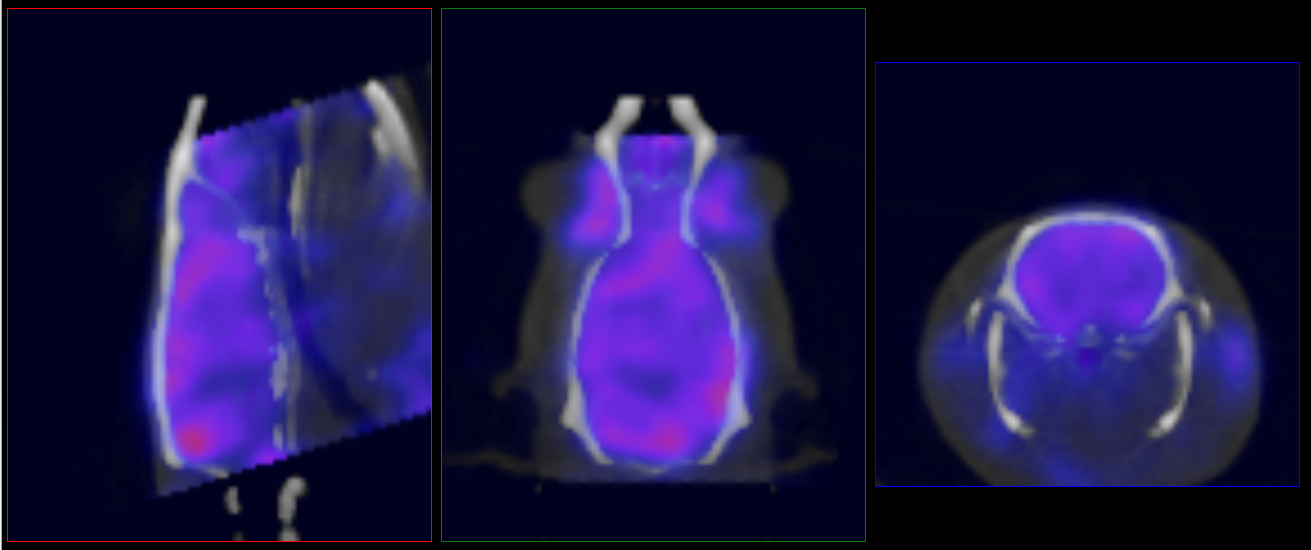
Once satisfied with the registration, unload the atlas anatomical reference and rerun the tool using the “Import 3D ROI” option under the advanced settings tab.